Database
Database Package
[Next: Vertebra Segmentation]
.v1 Database Package (Release date: 05-Oct-2015)
The .v1 database is packed into the xVertSeg.v1.zip archive file and is the first compilation of the database package, released on 05-Oct-2015.
The database package is composed of the following folders and files:
xVertSeg.v1/This is the root folder of the .v1 database package.Data1/This folder contains CT lumbar spine images with corresponding reference vertebra segmentation masks and fracture classification scores. The data in this folder should be therefore used for method research and training purposes.images/This folder contains CT lumbar spine images.masks/This folder contains reference vertebra segmentation masks that correspond to spine images in theData1/images/folder.scores/This folder contains reference fracture classification scores that correspond to spine images in theData1/images/folder and are located in the filescores.csv.
Data2/This folder contains CT lumbar spine images, for which corresponding reference vertebra segmentation masks and fracture classification scores are known only to challenge organizers. The data in this folder will be therefore used for method validation purposes.Matlab/This folder contains the Matlab support files.Results1/This folder has to be populated by participants and should contain results obtained for each spine image in theData1/folder.masks/Empty folder for vertebra segmentation results in the form of vertebra segmentation masks, preferably in MHD formatted files.scores/Empty folder for fracture classification results in the form of scores, preferably in a CSV formatted file.
Results2/This folder has to be populated by participants and should contain results obtained for each spine image in theData2/folder.masks/Empty folder for vertebra segmentation results in the form of vertebra segmentation masks, preferably in MHD formatted files.scores/Empty folder for fracture classification results in the form of scores, preferably in a CSV formatted file.
Spine Images
[Previous: Database Package] [Next: Vertebra Segmentation]
.v1 Spine Images (Release date: 05-Oct-2015)
The .v1 database contains 25 CT lumbar spine images including non-fractured vertebrae and vertebrae with fractures of different morphological grades and cases. The images are provided the MetaImage (MHD) format, a medicine image format used in the Insight Segmentation and Registration Toolkit (ITK) and other medical visualization software, according to which each image is represented by a header file (*.mhd) and a data file (*.raw) forming a pair.
- Data1
- For the 15 images grouped under “Data1” that are located in the
Data1/images/folder and named sequentially fromimage001.mhd/.rawtoimage015.mhd/.raw, corresponding vertebra segmentation masks and fracture classification scores are provided. These images should be therefore used for research and/or training purposes. - Data2
- For the 10 images grouped under “Data2” that are located in the
Data2/images/folder and named sequentially fromimage016.mhd/.rawtoimage025.mhd/.raw, corresponding vertebra segmentation masks and fracture classification scores are known to challenge organizers only. These images should be therefore used for testing purposes, and will be used by organizers for performance evaluation.
| .v1 Spine Image Database | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ID | Spine image | Precision (bits) |
Size (pixels) | Voxel size (mm/pixel) | Subject | |||||||
| Name | Group | X | Y | Z | ΔX | ΔY | ΔZ | Gender | Age (yrs) | |||
| .v1 Spine Image Database | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ID | Spine image | P (bits) |
Size (pixels) | Voxel size (mm/pixel) | Subject | |||||||
| Name | Group | X | Y | Z | ΔX | ΔY | ΔZ | G | A (yrs) | |||
Vertebra Segmentation
[Previous: Spine Images] [Next: Fracture Classification]
.v1 Vertebra Segmentation (Release date: 05-Oct-2015)
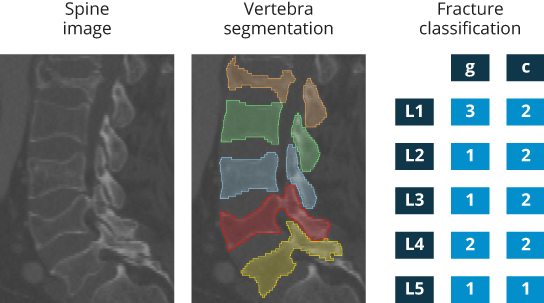
Vertebra segmentation is provided in the form of volume masks that assign each image pixel to a specified vertebral level or to the background, and was defined through the consensus of two observers (medical imaging scientists).
- Data1
- For the 15 images grouped under “Data1”, reference vertebra segmentation masks are located in the
Data1/masks/folder and named sequentially frommask001.mhd/.rawtomask015.mhd/.raw. Each lumbar vertebral level from L1 to L5 is assigned a unique mask value \(m\): $$\begin{split} L1:& \quad m = 200 \\[0.5em] L2:& \quad m = 210 \\[0.5em] L3:& \quad m = 220 \\[0.5em] L4:& \quad m = 230 \\[0.5em] L5:& \quad m = 240 \end{split}$$ with the value \(m=0\) representing the background. - Data2
- For the 10 images grouped under “Data2”, reference vertebra segmentation masks are known to the challenge organizers only. The task of participants is therefore to create vertebra segmentation masks for these images, where each lumbar vertebral level from L1 to L5 is assigned the following mask values \(m\): $$\begin{split} L1:& \quad 195 \leq m \leq 205 \\[0.5em] L2:& \quad 205 \leq m \leq 215 \\[0.5em] L3:& \quad 215 \leq m \leq 225 \\[0.5em] L4:& \quad 225 \leq m \leq 235 \\[0.5em] L5:& \quad 235 \leq m \leq 245 \end{split}$$ with the value \(m=0\) representing the background. Overlapping mask values will be considered to belong to the corresponding two vertebrae. For example, if a pixel is assigned a mask value of \(m=215\), it will be assumed that that pixel belongs both to L2 vertebra and to L3 vertebra.
Fracture Classification
[Previous: Vertebra Segmentation] [Next: Matlab Support]
.v1 Fracture Classification (Release date: 05-Oct-2015)
Fracture classification is provided in the form of scores that assign each vertebral level to a specified morphological grade and case of vertebral fractures by following the scoring scheme of Genant et al. [1], and was defined through the consensus of two observers (radiologists).
- Data1
- For the 15 images grouped under “Data1”, reference fracture classification scores are located in the
Data1/scores/folder in the comma-separated value (CSV) formatted filescores.csv. By skipping the first two lines and the first tabulated columns (which make the file readable by spreadsheet applications such as Microsoft Excel), the resulting values form a matrix where each row matches a spine image in the ascending order fromimage001toimage015, and values in each row follow vertebral levels in the ascending order from L1 to L5 and sequentially correspond to scores \(S=(g,c)\), where \(g\) is the grade and \(c\) is the case of vertebral fractures, in the form ofgL1,cL1,gL2,cL2,gL3,cL3,gL4,cL4,gL5,cL5. For example, row number 7 is equal toimage007,3,2,1,2,1,2,2,2,1,1, which means that for spine imageimage007, the score for vertebral levels L1 is \(S_{L1}=(g_{L1},c_{L1})=(3,2)\) representing a severe (3) biconcave (2) fracture, the score for vertebral level L2 is \(S_{L2}=(g_{L2},c_{L2})=(1,2)\) representing a mild (1) biconcave (2) fracture, the score for vertebral level L3 is again \(S_{L3}=(g_{L3},c_{L3})=(1,2)\) representing a mild (1) biconcave (2) fracture, the score for vertebral level L4 is \(S_{L4}=(g_{L4},c_{L4})=(2,2)\) representing a moderate (2) biconcave (2) fracture, and the score for vertebral level L5 is \(S_{L5}=(g_{L5},c_{L5})=(1,1)\) representing a mild (1) wedge (1) fracture. - Data2
- For the 10 images grouped under “Data2”, reference fracture classification scores are known to challenge organizers only. The task of participants is therefore to generate fracture classification scores for these images.
| .v1 Reference Fracture Classification | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| ID | Spine image | L1 | L2 | L3 | L4 | L5 | |||||
| Name | Grade | Case | Grade | Case | Grade | Case | Grade | Case | Grade | Case | |
| .v1 Reference Fracture Classification | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| ID | Spine image | L1 | L2 | L3 | L4 | L5 | |||||
| Name | g | c | g | c | g | c | g | c | g | c | |
Matlab Support
[Previous: Fracture Classification] [Next: Download]
The Matlab Support is located in the Matlab/ folder of the database package and consists of Matlab functions that participants are encouraged to use to load and save data, and to evaluate the performance of their methods.
loadVolume.m
A single volume (spine image or segmentation mask) in the MHD format can be loaded by function:
[H, I] = loadVolume(fileName)
The input argument fileName is the filename of the target MHD file (complete with path and *.mhd extension, *.raw extension or filename only without extension). The output argument H is a structure containing volume information, and the output argument I is a matrix containing volume image data, with matrix columns representing the left-to-right (\(x\)) anatomical direction, matrix rows representing the anterior-to-posterior (\(y\)) anatomical direction, and matrix layers representing the cranial-to-caudal (\(z\)) anatomical direction.
saveVolume.m
A single volume (spine image or segmentation mask) can be saved to the the MHD format by function:
saveVolume(H, I, fileName, overWrite)
The input argument H is a structure containing volume information, the input argument I is a matrix containing volume image data, the input argument fileName is the filename of the target MHD file (complete with path and *.mhd extension, *.raw extension or filename only without extension), and the input argument overWrite defines whether an existing file can be overwritten (default value 0, force value 1 to allow overwriting).
evalSegmentation.m
Evaluation metrics for vertebra segmentation performance can be computed by function:
[DSC, MSSD] = evalSegmentation(segM, refM, voxDim)
Input arguments segM and refM represent, respectively, the segmented and the corresponding reference volume masks in the form of matrices (values equal to 0 represent the background, and values different from represent vertebral levels), and the input argument voxDim = [xDim, yDim, zDim] is the vector of voxel dimensions (mm/pixel). The output arguments represent vertebra segmentation evaluation metrics, where DSC represents the DSC and MSSD represents the MSSD (in millimeters). Note that both output arguments are vectors, where each element corresponds to a vertebral level in the given volume masks. The function requires the file bwdistsc.m located in the same directory, and that the Image Processing Toolbox is installed with Matlab.
loadScores.m
Vertebra fracture scores can be loaded by function:
S = loadScores(fileName)
The input argument fileName is the filename of the target CSV file (complete with path and extension). The output argument is a matrix containing vertebra fracture scores, with matrix rows representing spine images, matrix columns representing vertebral levels, and matrix layers representing fracture grade (layer 1) and fracture case (layer 2).
saveScores.m
Vertebra fracture scores can be saved by function:
saveScores(S, fileName, overWrite)
The input argument S is a matrix containing vertebra fracture scores, the input argument fileName is the filename of the target CSV file (complete with path and extension), and the input argument overWrite defines whether an existing file can be overwritten (default value 0, force value 1 to allow overwriting).
evalClassification.m
Evaluation metrics for fracture classification performance can be computed by function:
[MSPP, F] = evalClassification(detS, refS)
The input arguments detS and refS represent, respectively, matrices of the detected and reference scores, with matrix rows representing spine images, matrix columns representing vertebral levels, and matrix layers representing fracture grade (layer 1) and fracture case (layer 2). The output arguments represent fracture classification evaluation metrics, where MSPP represents the MSSP and F represents the F-score.
Download
[Previous: Matlab Support]
Although contact information is not mandatory in order to download the database, by downloading you confirm and agree that:
- the owner of the downloaded material is the Laboratory of Imaging Technologies (University of Ljubljana, Faculty of Electrical Engineering, Slovenia),
- the downloaded material will be primarily used for purposes related to the challenge, if used for other purposes, written permission has to be obtained from the Laboratory of Imaging Technologies (University of Ljubljana, Faculty of Electrical Engineering, Slovenia),
- the downloaded material will not be used for commercial purposes, and will not be modified, copied and/or distributed in any form.

